Native Proteins
Characterizing Native Protein Structures in Cultural Heritage Objects
About the project
Native mass spectrometry techniques, unlike bottom-up and top-down methodologies, are capable of directly measuring multimeric protein complexes while retaining their non-covalent interactions.1–4 The capability to measure protein structures that closely resemble their solution counterparts enables new lines of research on how changes to the proteins structure as a function of changes to sequence, post translational modifications (PTM’s), or forced degradation. Basically, instead of breaking the protein of interest into fragments for analysis, native mass spectrometry allows you to look at the whole protein and all of its chemical changes. This tells you a lot about how a protein’s structure changes with ageing and interactions with mineral pigments or other organic materials (oils, polysaccharides, resints). To accurately characterize ancient protein structures found in objects of cultural heritage, such as paintings, it is critical to first understand the changes in protein structures induced from the chemical processes they undergo.

Hierarchy of protein structures measured by mass spectrometry. Native proteins retain their noncovalent interactions and complexes. Top-down methods fragment proteins, either native or intact, to obtain large peptide sequence information. Middle-down and bottom-up start with
already broken apart protein structure to measure smaller peptide sequence information.

nESI emitter under microscope. The orifice at the tip of the emitter for nano electrospray is often 1 – 2 micrometers large and under and electric potential of 0.5 – 1.5 kV.
What are native proteins?
With appropriate nano electrospray ionization (nESI) conditions, it is possible to transfer proteins and protein complexes to the gas-phase while retaining their non-covalent interactions.5 These so-called “native” protein structures closely resemble their solution-phase counterparts in the absence of bulk solvent.3 Additionally, native methodologies are advantageous because they used to directly measure the changes to a protein’s 3D conformational structure, protein-complex stoichiometry, and changes to stability.
Traditional proteomic techniques such as bottom-up and top-down analyze protein sequences rather than their intact structure. Such techniques have found relatively recent success in the CH field and have been applied towards the authentication of glues6, validating milk as a secret ingredient in Gainsborough’s drawings7, and the analysis of collagen in palaeontologic fossils8,9.While obtaining full sequence coverage is possible for abundantly available purified proteins, it is challenging to achieve similar sequence coverage for proteins that are sample-restricted like those from CH objects.
Additionally, losing the proteins’ native structure through denaturation or proteolytic digestion results in the loss of a wealth of information content relevant to the proteins structural conformations, changes in stability, and network of interactions. By studying the nativity of protein structures we aim to fill the gaps in our understanding of native protein structures in CH, and in doing so assist future conservation efforts to avoid further protein degradation that may affect an objects’ appearance.
Why native proteins?
Through spectroscopic methods such as Fourier transform infer-red (FTIR) and Raman sepctroscopy, or immuno-assays such as enzyme-linked immunosorbent assay ( ELISA), it is possible to find differences between protein controls and ancient proteins found in paintings. However, it is not currently known whether the changes that protein structures undergo are solely due to the stress they experience over the course of their lifetimes embedded in the paint, or if this is stress induced during the drying process of the paint layer. Previous studies have indicated that secondary structure of globular proteins is minimally affected by dehydration, but that intrinsically disordered proteins become more structured during similar experiments10. However, other reports have contrasted this finding indicating dehydration induces significant changes to structural conformation, but these effects can be mitigated in the presence of stabilizers such as sucrose11. There is a need to answer questions revolving around whether protein structures embedded in paintings are inherently perturbed, or if the heterogeneity of materials and molecular crowding stabilize structures during the drying process. In the case where both are true, there are deeper questions on what paint conditions (i.e. composition of pigments, binders, or additives) perturb or preserve protein structure, and if such knowledge can be leveraged to predict protein structure found in other paintings of similar composition Answering these questions regarding changes in protein structure and conformation will have rippling affects for the future interpretation of ancient proteins studied for cultural heritage.
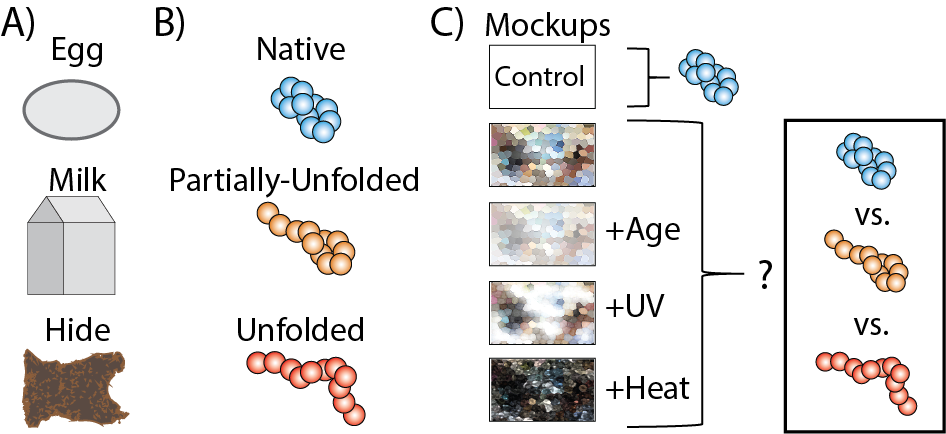
(A) protein origin and class of proteins most commonly found in CH objects and paintings. (B) A series of paint mockups to represent different stress states embedded proteins would experience. The resultant IM-MS and CIU data will be used to evaluate change in protein structure.
Further Reading
(1) Light-Wahl, K. J.; Winger, B. E.; Smith, R. D. Observation of the Multimeric Forms of Concanavalin A by Electrospray Ionization Mass Spectrometry. J. Am. Chem. Soc. 1993, 115 (13), 5869–5870.
(2) Light-Wahl, K. J.; Schwartz, B. L.; Smith, R. D. Observation of the Noncovalent Quaternary Associations of Proteins by Electrospray Ionization Mass Spectrometry. J. Am. Chem. Soc. 1994, 116 (12), 5271–5278.
(3) Ruotolo, B. T.; Giles, K.; Campuzano, I.; Sandercock, A. M.; Bateman, R. H.; Robinson, C. V. Biochemistry: Evidence for Macromolecular Protein Rings in the Absence of Bulk Water. Science (80-. ). 2005, 310 (5754), 1658–1661.
(4) Park, A. Y.; Jergic, S.; Politis, A.; Ruotolo, B. T.; Hirshberg, D.; Jessop, L. L.; Beck, J. L.; Barsky, D.; O’Donnell, M.; Dixon, N. E.; Robinson, C. V. A Single Subunit Directs the Assembly of the Escherichia Coli DNA Sliding Clamp Loader. Structure 2010, 18 (3), 285–292.
(5) Fenn, J.; Mann, M.; Meng, C.; Wong, S.; Whitehouse, C. Electrospray Ionization for Mass Spectrometry of Large Biomolecules. Science (80-. ). 1989, 246 (4926), 64–71.
(6) Dallongeville, S.; Richter, M.; Schäfer, S.; Kühlenthal, M.; Garnier, N.; Rolando, C.; Tokarski, C. Proteomics Applied to the Authentication of Fish Glue: Application to a 17th Century Artwork Sample. Analyst 2013, 138 (18), 5357–5364.
(7) Pozzi, F.; Arslanoglu, J.; Galluzzi, F.; Tokarski, C.; Snyder, R. Mixing, Dipping, and Fixing: The Experimental Drawing Techniques of Thomas Gainsborough. Herit. Sci. 2020, 8 (1), 1–15.
(8) Demarchi, B. Ancient Protein Sequences. Amin. Acids Proteins Foss. Biominer. 2020, 113–126.
(9) Buckley, M.; Larkin, N.; Collins, M. Mammoth and Mastodon Collagen Sequences; Survival and Utility. Geochim. Cosmochim. Acta 2011, 75 (7), 2007–2016.
(10) Yoneda, J. S.; Miles, A. J.; Araujo, A. P. U.; Wallace, B. A. Differential Dehydration Effects on Globular Proteins and Intrinsically Disordered Proteins during Film Formation. Protein Sci. 2017, 26 (4), 718–726.
(11) Prestrelski, S. J.; Tedeschi, N.; Arakawa, T.; Carpenter, J. F. Dehydration-Induced Conformational Transitions in Proteins and Their Inhibition by Stabilizers. Biophys. J. 1993, 65 (2), 661–671.
Research performed in collaboration with…
This page is based upon work supported by the National Science Foundation Mathematical and Physical Sciences divisions ASCEND program under award number CHE-2138107.
Any opinions, findings, and conclusions or recommendations expressed in this material are those of the author(s) and do not necessarily reflect the views of the National Science Foundation.









