Ivory and Bone
Species Identification of Ancient Ivory and Bone (Egyptian and Medieval) Using Micro-sampling and Proteomics
What is the question?
There are many objects from the Metropolitan Museum of Art’s Egyptian Department and European Sculpture and Decorative Arts Department which are made of bone and ivory. Typically, bone and ivory can be differentiated microscopically by its porosity (vasculature). In the case of ivory, the presence of growth lines can also indicate the species of origin. However, in the case of highly carved and polished objects, it is not known whether these objects are made from bone or ivory, or from what species they originate. In this project we want to perform the species identification of these precious objects in order to better understand the conservation needs of the object, and to shed light on the context in which these objects were created.
Some of the bone and ivory objects in the Metropolitan Museum of Art’s Egyptian collection that have been sampled.
What is ivory?
Ivory originates from the tusks of animals such as elephants, narwhals and hippos, which are essentially elongated teeth. Ivory is a popular sculpting material in many cultures because of its hardness, and because it is possible to polish it to a shiny surface without the need of additional resins and varnishes.1
Like bones and teeth, ivory is a mineralised tissue. These tissues are mainly composed of the mineral hydroxyapatite, Ca5(PO4)3(OH), together with water and proteins in varying proportions.2 Of the proteins present in these tissues, more than 90% is composed of collagens, a group of structural proteins. It is these proteins on which we will focus during this project.
Minimally Invasive Sampling
In many studies of ancient proteins in bones and teeth, samples are taken using specialised drills in order to remove sufficient sample to have conclusive results.3 However, it is not possible to apply such techniques to precious museum objects, and so we have used minimally invasive sampling techniques, specifically the use of a microgrit polishing paper, and developed sample preparation methods for small amounts of material, to overcome this hurdle.4,5

Using a microgrit to sample immeasurably small amounts of material from the surface of (a) bone and (b) teeth, which were used as an ivory model.
Ancient proteins for species identification
Since DNA encodes the amino acid sequence of proteins, proteins, such as collagens, are a source of taxonomic information. Additionally, proteins have far stronger interactions with the mineral matrix than DNA, and so are preserved in mineralised tissues for far longer periods of time.6 This is particularly true for structural proteins such as collagens. The sequence of amino acids in collagens vary slightly between different species, therefore, using proteomics, we can detect these differences and identify the species of origin of the objects. However, the amino acid sequence of such structural proteins is highly conserved, so in order to be able to detect these small differences in the amino acid sequence, a sample preparation method with minimal loss, and a highly sensitive analytical method needed to be developed.
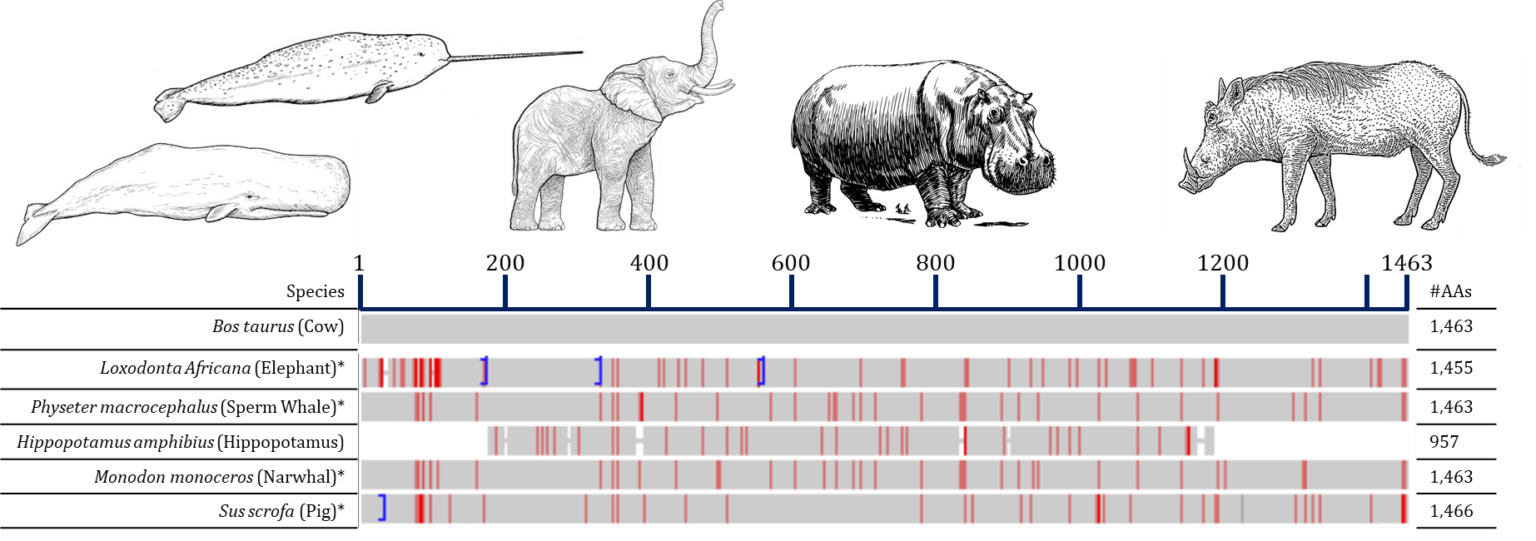
Sequence alignment of collagen alpha-1(I) of some of the species of interest in this project, performed using the COBLAT algorithm, available on NCBI, All species were compared against Bos taurus, red lines indicate where residues differ between the species and Bos taurus, while blue lines indicate that some amino acids are not present.
This project is particularly challenging for two main reasons. Firstly, as discussed above there is not a lot of variation between the collagens of different species, especially if they are closely related. Therefore, our sample preparation procedures must be optimised in order to detect the highest sequence coverage of collagen possible, in order to increase our chances of detecting species specific parts of the protein. Secondly, many species from which ivory originates, for example elephants and narwhals, have not had their proteome fully sequenced. This means that a traditional proteomics experiment involving a database search will be challenging, and that the proteins much be sequenced from scratch, de novo. In addition to this, the use of ivories with a known origin will be used in order to build our own home-made databases based on proteomics data rather than DNA-derived sequences, in order to have more confident species identifications.
Further Reading
1 Coutu, A. N., Whitelaw, G., le Roux, P. & Sealy, J. Earliest Evidence for the Ivory Trade in Southern Africa: Isotopic and ZooMS Analysis of Seventh–Tenth Century ad Ivory from KwaZulu-Natal. African Archaeol. Rev. 33, 411–435 (2016).
2 Cerling, T. E. et al. Radiocarbon dating of seized ivory confirms rapid decline in African elephant populations and provides insight into illegal trade. Proc. Natl. Acad. Sci. U. S. A. 113, 13330–13335 (2016).
3 Welker, F. et al. The dental proteome of Homo antecessor. Nature 580, 235–238 (2020).
4 Pozzi, F., Arslanoglu, J., Galluzzi, F., Tokarski, C. & Snyder, R. Mixing, dipping, and fixing: the experimental drawing techniques of Thomas Gainsborough. Herit. Sci. 8, 1–14 (2020).
5 Dallongeville, S., Garnier, N., Rolando, C. & Tokarski, C. Proteins in Art, Archaeology, and Paleontology: From Detection to Identification. Chem. Rev. 116, 2–79 (2016).
6 Demarchi, B. et al. Protein sequences bound to mineral surfaces persist into deep time. Elife 5, (2016).
Research performed in collaboration with…
This project has received funding from the European Union’s Horizon 2020 research and innovation programme under the Marie Sklodowska-Curie grant agreement No.861389













